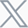跳转地址1Advancing Novel Breast Cancer Drug Development with HER2-ADC Resistance Models
According to data from the World Health Organization (WHO), breast cancer is the second most commonly diagnosed cancer globally, after lung cancer, and remains the leading cause of cancer incidence and mortality among women.
The evolution of HER2-targeted therapy has progressed through multiple stages. In 2019, the antibody-drug conjugate (ADC) T-DXd (Trastuzumab Deruxtecan, Enhertu) was approved, offering a new strategy to overcome resistance to the previous generation ADC drug, T-DM1. The cytotoxic payload, DXd, carried by this drug is membrane-permeable. This property allows it not only to precisely target HER2-high-expressing cancer cells but also to eliminate adjacent HER2-negative tumor cells through the "bystander effect," resulting in superior efficacy. However, as clinical use expands, acquired resistance inevitably emerges in some patients. Consequently, establishing reliable preclinical resistance models has become crucial for advancing subsequent research.
Current strategies for constructing resistance models primarily fall into four categories (Figure 1):
Patient-Derived Xenograft (PDX) Models: These models transplant resistant tumor tissue from patients into immunodeficient mice. They effectively preserve tumor heterogeneity and are suitable for individualized therapy research, but face challenges such as long modeling cycles and limited reproducibility.
In Vivo Induced Resistance: This method simulates the evolution of resistance by inoculating sensitive tumor cells into mice and repeatedly administering the drug.
It closely mirrors the real physiological environment, but the overall cycle remains relatively long.In Vitro Induced Resistance: This approach involves gradually increasing drug concentration in the cell culture medium to select for resistant cell lines under drug pressure. It is convenient and has a short cycle, but carries the risk of insufficient correlation between in vitro and in vivo results.
Gene-Edited Resistance Models: This technique utilizes gene-editing technology to introduce specific resistance mutations, allowing for the precise study of single mechanisms. However, it struggles to replicate the multi-gene, multi-pathway complexity of clinical resistance.